Geodata in Python: an example from Australian Covid-19 data
Code associated to this post: https://github.com/opsabarsec/Cleaning-and-EDA-Australian-COVID-19-data-
Australia 2020: the first COVID-19 wave just hit the country. Knowing where the virus is spreading faster, identifying clusters etc. are extremely important information to tackle pandemic. But such information is not so straightforward to obtain.
There are a few challenges :
- data may not be ‘‘clean’’ : who records testing results has loads of work, since testing structures have to be built from scratch as fast as possible. It is normal that the data will contain duplicates and misspellings.
- visualization needs to be clear even to a non-scientific public, this will also help to understand and apply precautions, distancing etc.
- graphs need to be daily updated.
1. Data cleaning
Deduplication as mentioned earlier is not straightforward. We cannot just apply the Python “dedupe” library to a test or patient ID. This would not include duplicates made by misspellings as in the example below (table 1)

This requires grouping fields with similarity criteria. One solution is to use a dedicated software to process data prior and feed them later to a Python script that will extract the relevant information. An open source software for this task is OpenRefine[1]. But in this case I decided to add this process directly in the pipeline using the library “fuzzywuzzy”[2] using the following code.
tested_containing_dupes = df1[feature_to_deduplicate] deduped_feature_tested = fuzzywuzzy.process.dedupe(tested_containing_dupes) df1_deduped = df1.loc[df1[feature_to_deduplicate].isin(deduped_feature_tested)]
This package uses as criterium the Levenshtein distance: the amount of duplicates removed with this method was as high as 31.25% of the data.
After deduplication, outliers and unreasonable values (say, negative age or 120 years old patients) need also to be detected. Values can be then replaced by a value obtained birthdate if the latter is reasonable.
To obtain clean geographical information, when the state name was misspelled the correct value was obtained with some feature engineering, using the postcodes.
2. The Chloropleth maps
A choropleth map is a map of a geographic area, in which different regions are represented by a color or pattern based on an aggregated attribute of that particular subregion (Australian states in this case). Choropleth maps are one of the most effective methods to visualize geographic data. In Python they can be obtained using the “folium” library[3].
But to apply it to a specific part of the world such as Australia, a shapefile is needed. Shapefiles are data structures that contain information about different geographic regions. They contain the geometric representation of the regions, which we will need to map them. Besides, the shapefiles optionally contain some additional metadata like name of regions. These can be found for several countries, normally in GitHub repositories and imported using the following code
from urllib.request import urlopen
import json
with urlopen('https://raw.githubusercontent.com/rowanhogan/australian-states/master/states.geojson') as response:
state_geo = json.load(response)# Read the geojson data with Australia's state borders from githubCombining this with the clean data a self-explanatory map can be finally obtained.
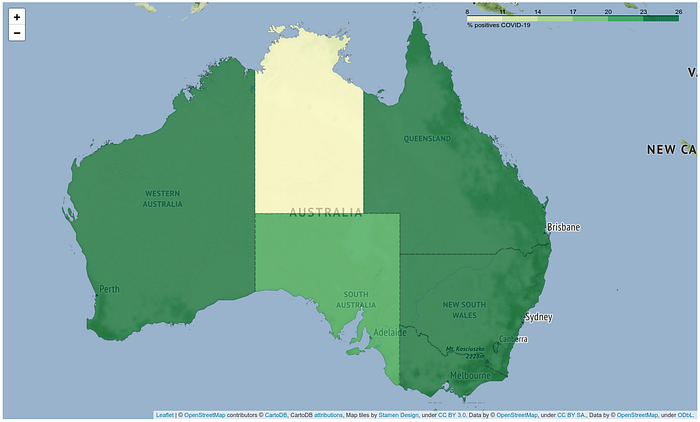
A small detail is that this will be saved as html page. But a png or jpg image can be obtained from it using the following code so that it can be displayed also in GitHub[4].
import Imagemap_data = m._to_png(5)
mappa = Image.open(io.BytesIO(map_data))
mappa.save('images/cloropleth.png')
image3 = Image.open("images/cloropleth.png")
image3
Conclusions
- A cleaning pipeline was created by de-duplicating entries using phone number in a first pass, then a dedicated Python library, “fuzzywuzzy”.
- Chloropleth maps could be obtained and embedded into the code using the library “folium”. These can display efficiently the Covid-19 spreading.
References
2) https://pypi.org/project/fuzzywuzzy/
